Cellular microarray
Cellular microarray provides a platform to visualize and index cells amenable to high-throughput studies. We present a dielectrophoresis (DEP)-based cellular microarray chip integrated with a concentration gradient generator for cellular microarray applications. Positive DEP force was applied to pattern cells on collagen-coated planar interdigitated ring electrode (PIRE) array. Key features of the PIRE design include: (1) allowing most edges where the localized maximum in the electric field exists; (2) making the inner gap slightly smaller than the outer gap in causing the electric field strength near the center of a PIRE being generally stronger than that near the outer edge of the same PIRE. Results show the integrated concentration gradient generator provides concentrations with high repeatability. Further, human hepatocellular carcinoma cells, HepG2, adhered on a 6×6 PIRE array show that cells patterned within minutes with good uniformity (48±6 cells per PIRE). Cell viability test revealed healthy patterned cells after 24 h that were still confined to the collagen-coated PIREs. Quantification of fluorescence intensity of living cells shows an acceptable reproducibility of cell viability among PIREs (mean normalized intensity per PIRE was 1±0.138). This integrated chip would allow batch testing with different stimulus concentrations in a single chip for high-throughput cellular studies.The result also suggest that the PIRE array would benefit applications that require uniform cellular patterning with fewer cells, and improve both response and reproducibility of cell-based biosensors. The next step of this study is to utilize the chip for an in vitro assessment to determine efficacy of a particular chemotherapeutic regimen prior to actual treatment.
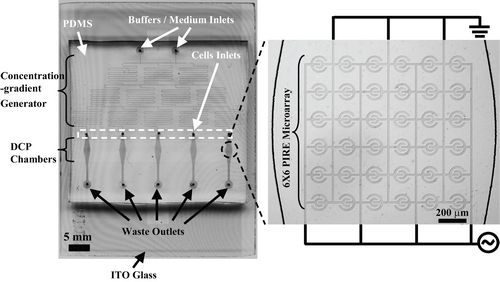
Figure 1. The integrated microfluidic cellular microarray chip. The chip is consisted of two main parts: planar interdigitated ring electrode (PIRE) of five 6×6 arrays in downstream of a PDMS concentration gradient generator. The PIRE arrays are designed to uniformly pattern cells via dieletrophoretic cell patterning (DCP). The concentration gradient generator can provide stable concentrations of stimulus flowing over patterned cells.
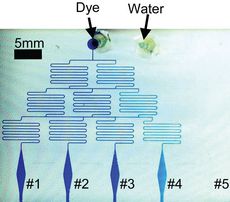
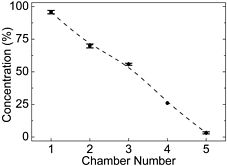
Figure 2. The PDMS concentration gradient generator. (a) Top-view of the generator. Two streams entered from top inlets at the same flow rate of 5μL/min and mixed as they flowed downstream. (b) Measured data (mean of three measurements) showed a near-linear concentration gradient was achieved.
.jpg)
Figure 3. Dielectrophoretic cell patterning (DCP) on a 6×6 PIRE array with applied voltage of 5Vpp at 5MHz. (a) The patterned cells (HepG2) covered the entire PIRE, not only the edges (cell-seeding density was about 5×106 cells/mL). (b) Cell count on each PIRE shows good overall uniformity, 48±6 cells per PIRE. (c) The specific distribution of patterned cells in each row of electrode within the PIRE array. (d) The specific distribution of patterned cells in each column of electrode.
.jpg)
Figure 4. Cell viability of patterned HepG2 before and after 24 h. (a) Before incubation, the patterned cells were round and spherical in shape. (b) After 24 h incubation, cells became flat and polygonal in shape and were still confined to the area of each PIRE. (c) Stained with calcein AM, quantification of the fluorescence intensity of living cells shows mean normalized intensity per PIRE was 1±0.138. This suggests that the reproducibility of cell viability among PIREs is acceptable. (d) The composite of (b) and (c) reveals high cell viability by most cells being stained positively.